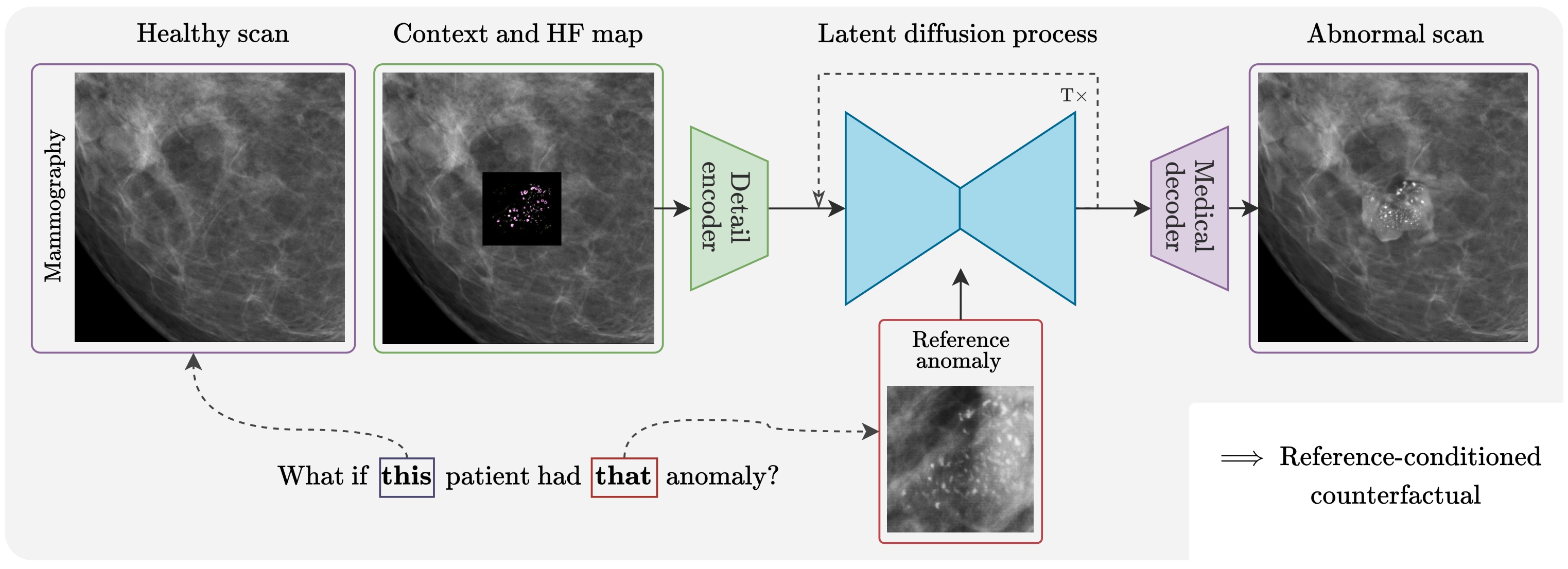
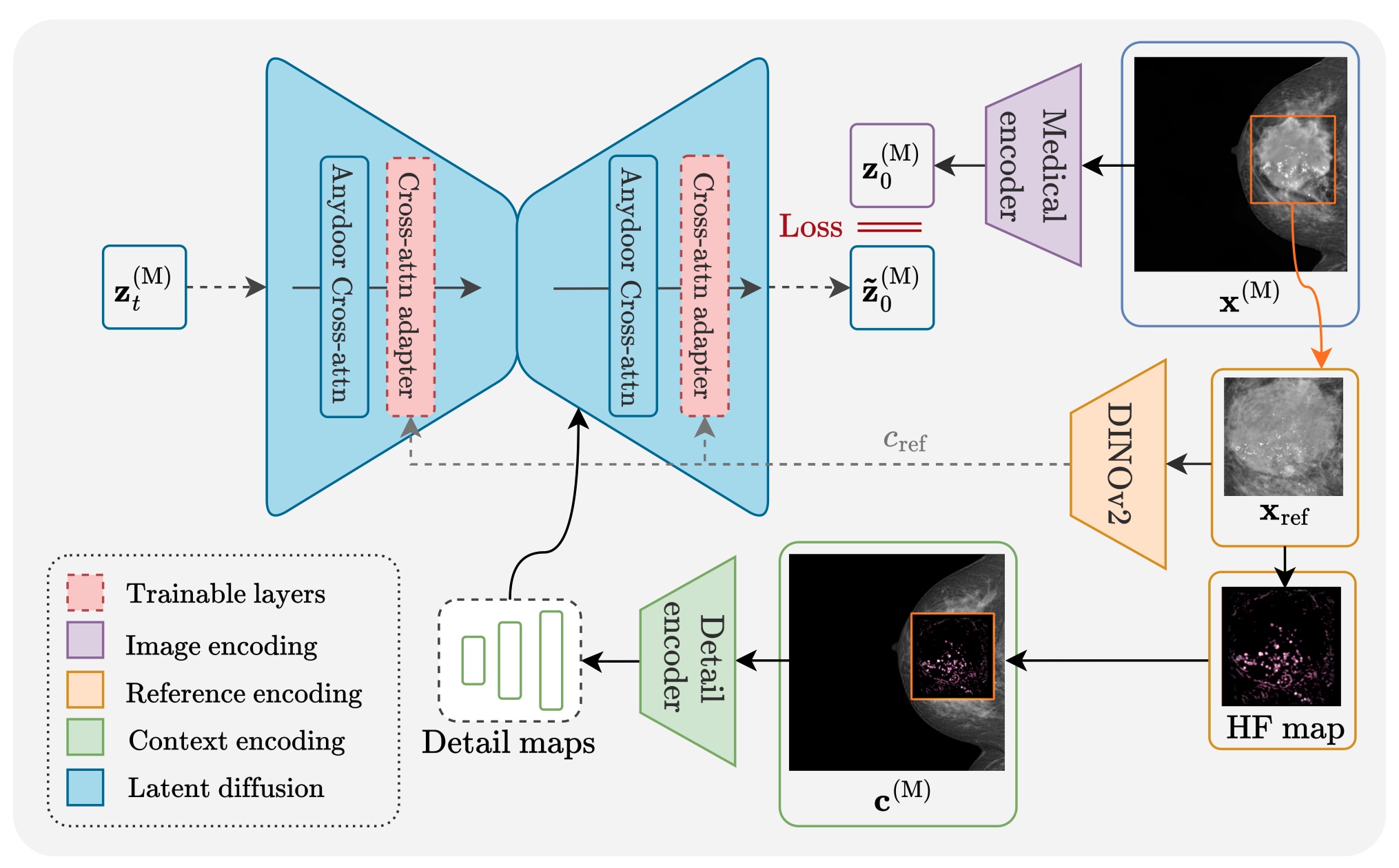
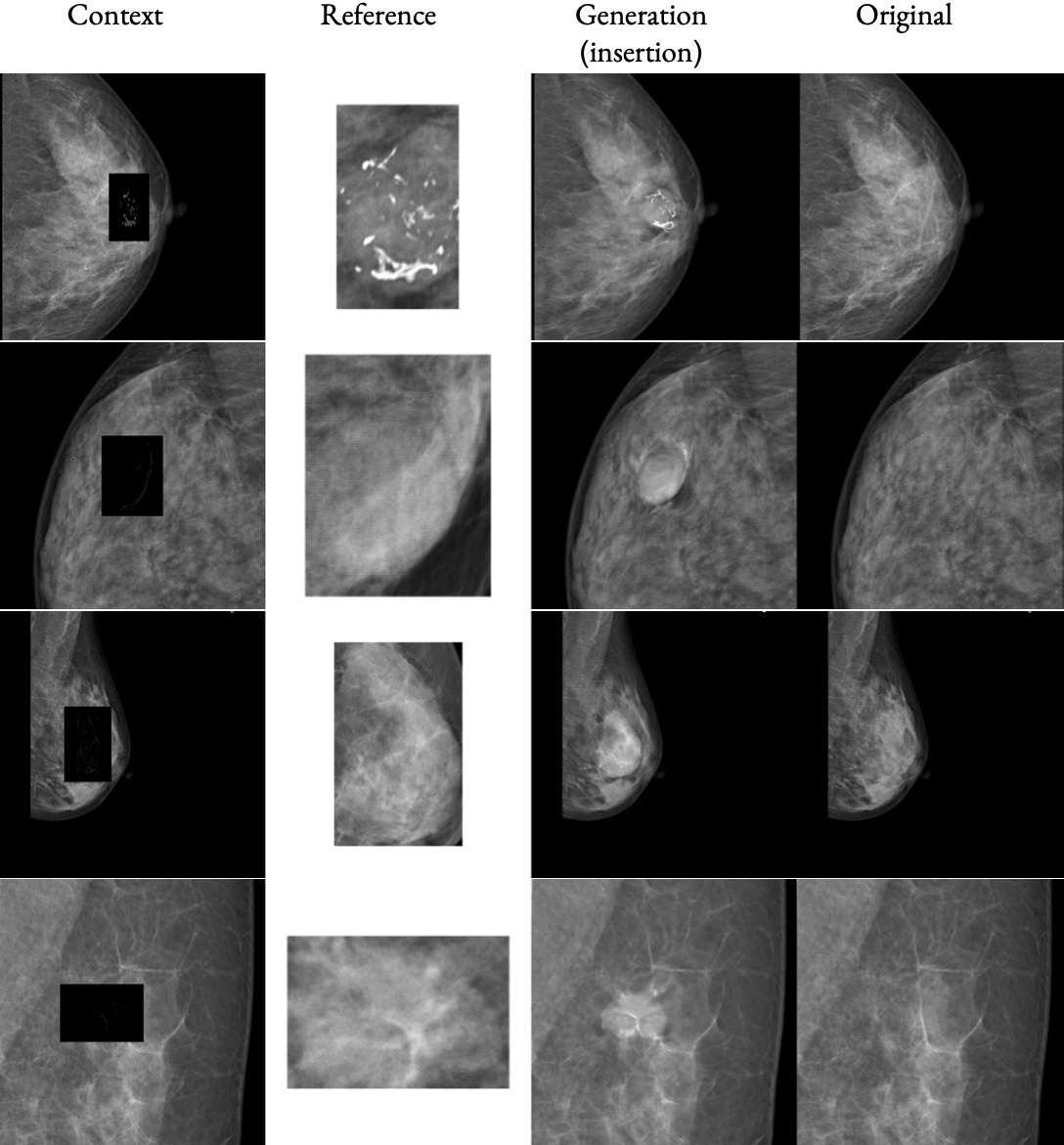
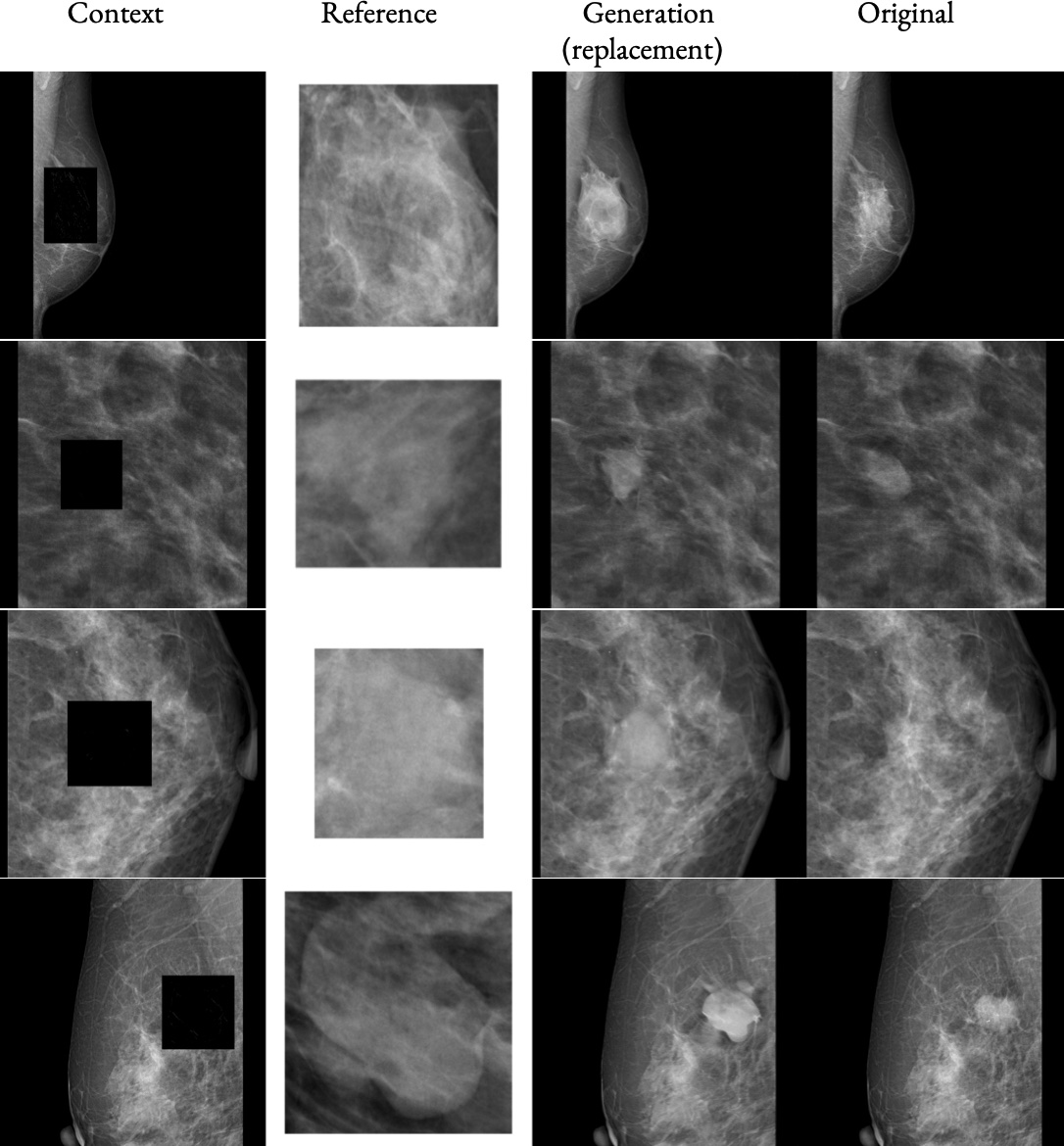
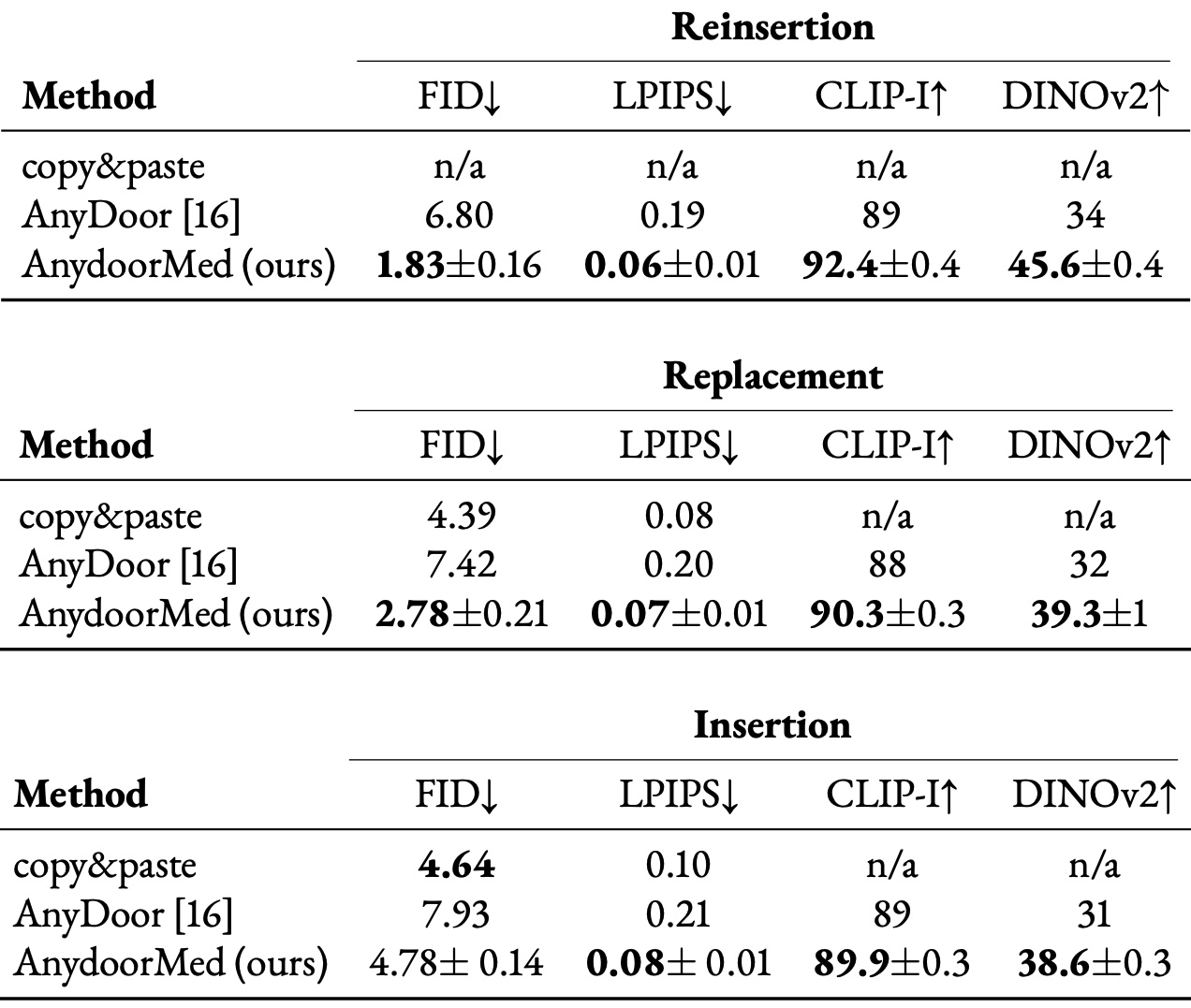
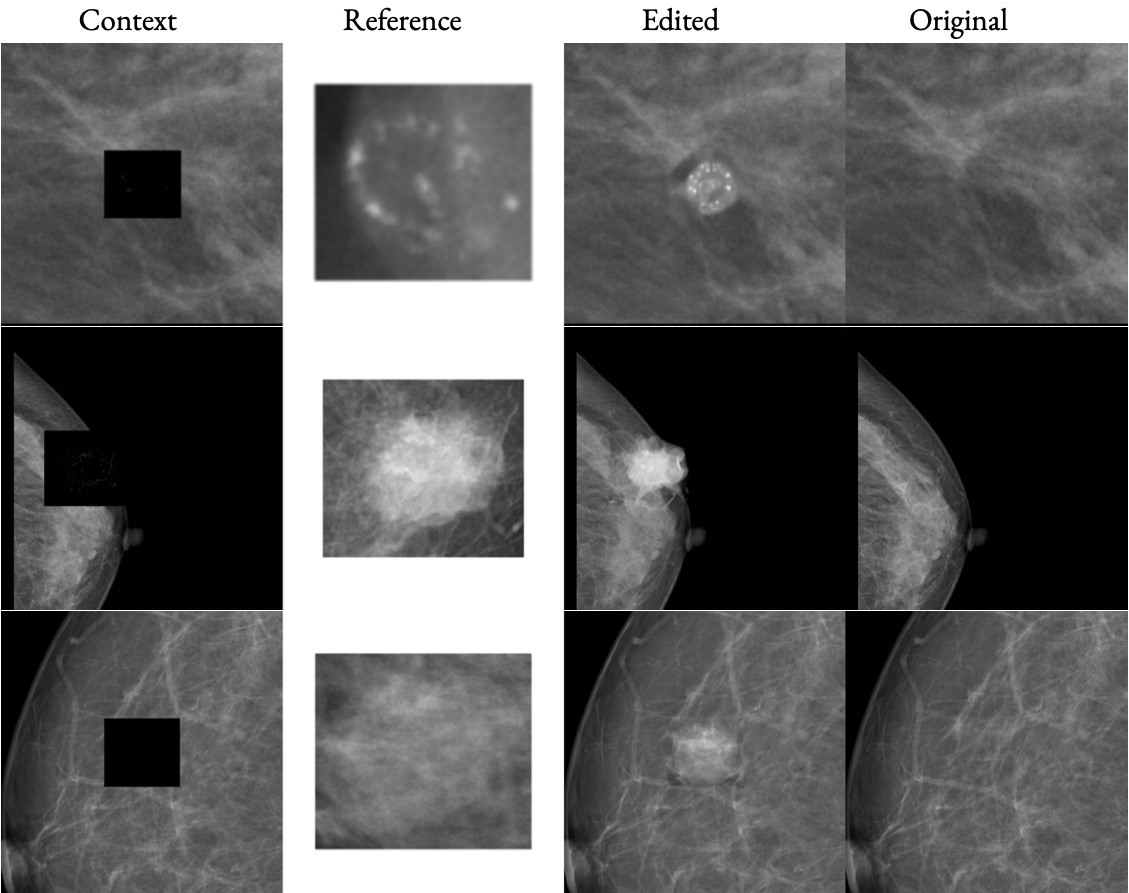
High-fidelity data is essential for developing reliable computer-aided diagnostics in medical imaging, yet clinical datasets are difficult to obtain and often suffer from severe class imbalance, particularly for rare pathologies such as malignant breast lesions. Synthetic data offers a promising avenue to mitigate these limitations by augmenting existing datasets with diverse and realistic counterfactual examples. To be clinically useful, generated data must respect anatomical constraints, preserve fine-grained tissue structures, and allow controlled insertion of abnormalities. While recent diffusion-based methods enable anomaly synthesis via text or segmentation masks, such conditioning often fails to capture subtle structural variations. In contrast, diffusion inpainting has shown promising results on natural images but remains underexplored in medical imaging. This works proposes AnydoorMed, a reference-guided inpainting method for mammography that transfers lesion characteristics from a source scan to a target location, blending them seamlessly with surrounding tissue, supporting the generation of realistic counterfactual examples.
@article{buburuzan2025reference,
title={Reference-Guided Diffusion Inpainting For Multimodal Counterfactual Generation},
author={Buburuzan, Alexandru},
journal={arXiv preprint arXiv:2507.23058},
year={2025}
}